 NA-MIC Project Weeks
NA-MIC Project Weeks
Back to Projects List
Real-time visualization for transcranial magnetic stimulation (TMS)
Key Investigators
- Loraine Franke (University of Massachusetts Boston)
- Jax Luo (BWH & Harvard Medical School)
- Yogesh Rathi (BWH & Harvard Medical School)
- Lipeng Ning (BWH & Harvard Medical School)
- Steve Pieper (Isomics, Inc.)
- Daniel Haehn (University of Massachusetts Boston)
Project Description
Transcranial magnetic stimulation is a nonivasive procedure used for treating depression with magnetic and electric fields to stimulate nerve cells. A TMS coil is slowly moved over the subject’s head suface to target certain areas in the brain. Our project aims to develop a deep-learning powered software for real-time E-Field prediction and a visualization of TMS within 3D Slicer.
Objective
Real-time visualization of an electric field (E-field) for transcranial magnetic stimulation (TMS) on the brain surface, visualization through an AR app (over browser).
Approach and Plan
What is done so far:
- We created a TMS module in Slicer mapping NifTi file onto brain mesh with 3D TMS coil that can be moved by the user.
- OpenIGTLinkIF is used to transfer data (E-Field from TMS) into 3D Slicer
- Connected 3DSlicer to the web browser using our newly implemented secure WebSocket from https://github.com/liampaulhus/slicerWebWSS-WIP
- Mobile device via WebXR connected and we can control the coil inside 3DSlicer.
- We have integrated a deep learning model (CNN) inside our SlicerTMS module. We receive real time updates of newly generated Nifti files via the OpenIGTlink Plugin. The current deep learning model predicts the TMS E-field. We visualized this field with the magnetic field of the coil in the correct position on the brain mesh.
- Beside the brain surface, we can visualize the E-Field on tractography fiber bundles. We have integrated the Fiber Bundle selection with an ROI attached to the TMS coil with the SlicerDMRI module.
Progress and Next Steps
- We improved the performance of the Fiber ROI selection by downsampling the fibers (see demo below).
- Fixed CUDA bug of the neural network model that generates the Nifti files to be visualized.
Illustrations
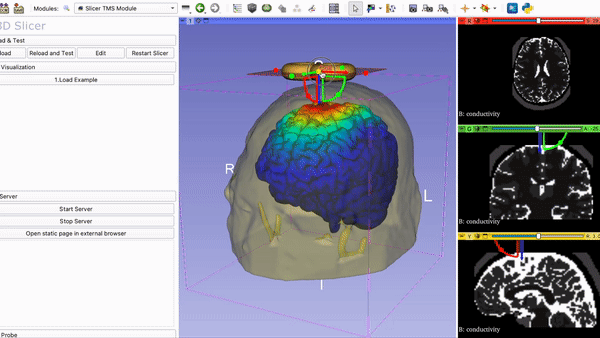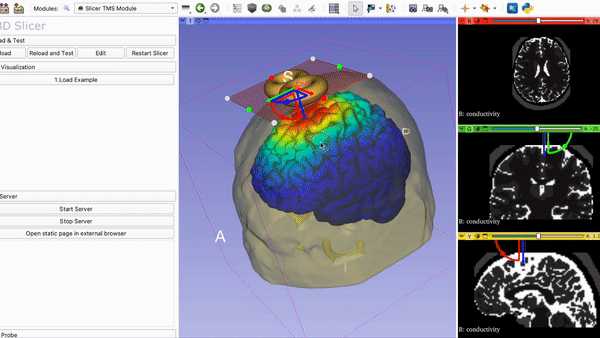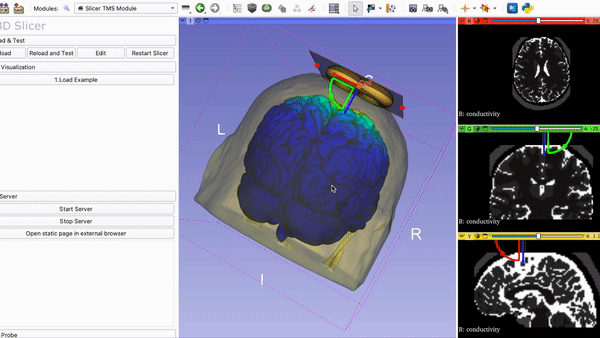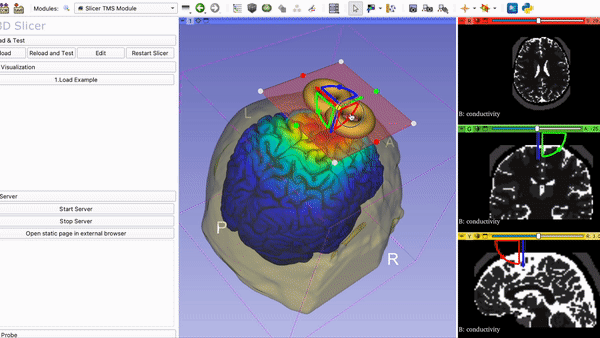
Current Visualization of the TMS Module in 3DSlicer with Coil and….
Mapping of E-field on tractography with ROI selection:

Mapping of E-field on brain surface:
Background and References
Infos for running WebXR:
Phones need a Depth sensor to run AR/VR. A list of supported devices can be found here: https://developers.google.com/ar/devices
On an Android Phone via USB:
- PlayStore: Download Google VR Services and Google AR Services App
- Update Chrome/Camera apps etc.
- On the phone: Enable Developer tools (https://developer.android.com/studio/debug/dev-options) and USB debugging (description here: https://developer.chrome.com/docs/devtools/remote-debugging/)
- Run chrome://inspect#devices in the browser on your computer and it should detect USB connected devices
For iPhone:
- Mozilla offers a WebXR Emulator that can be downloaded from the Apple Store for any iPhone and iPad: https://labs.mozilla.org/projects/webxr-viewer/
For the full SlicerTMS Module and instructions see our repository
Also see previous project week PW 37
<!– vtkProbeFilter: https://vtk.org/doc/nightly/html/classvtkProbeFilter.html Moving fiducials with CPYY: https://gist.github.com/pieper/f9da3e0a73c70981b48d0747132526d5
Measure rendering time in 3D Slicer:
- Getting renderer: https://slicer.readthedocs.io/en/latest/developer_guide/script_repository.html#access-vtk-views-renderers-and-cameras
- Then applying renderer.GetLastRenderTimeInSeconds()